Background
Throughout the first year in University, I took Object Oriented Programming class. While being a challenging class, it was also rewarding. It challenged me in various ways and allowed to develop a better understanding as well as higher competence in both Python programming and thinking.
For the final project in this class, we were tasked to create an Imputer, the task would explain an imputer as follows: “Most data science projects start by pre-processing a dataset to ensure the data is ready to use for its intended purpose. One of the tasks that a data scientist would typically complete during such a pre-processing phase is to replace missing data values in the dataset using a process known as imputation. “ In other words, we had to create a tool that would fill in the missing values with either a mean/mode/median.
Methodology
To achieve this, we were tasked to utilize Strategy Pattern. A strategy pattern involves separating classes in order to make them interchangeable, in this situation, the behaviors we wanted to change are between methods of imputation (mean/mode/median). Therefore I produced the code such as:
from abc import ABC, abstractmethod
class ImputerStrategy(ABC): # Interface for the Imputer class
@abstractmethod
def fit(self):
pass
@abstractmethod
def transform(self):
pass
class CalculateStrategy(ABC): # Interface for the Mean/Mode/Median class
@abstractmethod
def calculate(self):
pass
class AxisStrategy(ABC): # Interface for axis-specific strategies
@abstractmethod
def select(self, data):
passBrief explanation of the code:
-
ImputerStartegy - Is acting as an interface for the imputer class. This is where the we fit and transform the imputer itself
-
CalculateStrategy - Is the actual calculation startegy, at the moment it is the interface of the entire platform. This will make sure that we use the same methodology to calculate the mean/mode/median
-
AxisStrategy - This method was created to automatically sort by column and not row.
After establishing all the Interface classes, we can proceed to create the logic in each of the interfaces.
To make things easier for myself, i created first the logic for “Axis”, or in this case, so the code would only look through the columns instead of Rows.
Side note: For future self (if i find the need to use this program), i created a strategy as well as for rows.
class Axis0(AxisStrategy): #Axis0: Only corresponds to implementatiosn for Columns. NOT ROWS!!!!
def __init__(self,data):
self._data = data
def select(self, column_index):
return [row[column_index] for row in self._data if row[column_index] != "nan"] #Only extracts the columns, if the value is not nan.
class Axis1(AxisStrategy): #Axis1: Only corresponds to implentations for Rows. NOT NEEEDED !
def select(self, data):
pass In this code, the Axis0 will basically loop throughout the columns and Axis1 will go through the rows.
Furthermore, the logic for the calculation. As previously established interface, there will be 3 difference classes that inherit the logic from “CalculationStrategy” class. The code is as follows:
class Mean(CalculateStrategy):
def calculate(self,data):
return s.mean(data) #Calculates the mean and returns the data
class Mode(CalculateStrategy):
def calculate(self,data):
return s.mode(data) #Calulcates the mode and returns the data
class Median(CalculateStrategy):
def calculate(self,data):
return s.median(data) All the methods used were imported via “Statistics” library, which came in very convenient to create the necessary imputations.
And finally, to round it all together a seperate Imputer class was created. This class was responsible for storing all the previously established logic and produce an end result.
class Imputer:
def __init__(self,strategy:str="mean",axis:int=0):
self._strategy = strategy
self._axis = axis
self._ImputeValues = None #For storing the extracted data + applying one of the mean/mode/median calculations
self._axis_strategy = None
def fit(self, x, data):
self._axis_strategy = Axis0(data) if self._axis == 0 else Axis1() #This will determine weather it's for column or rows Axis 0 = Column/Axis1 = Row
column_data = self._axis_strategy.select(x)
#Strategy selection
if self._strategy == "mean":
calculator = Mean()
elif self._strategy == "mode":
calculator = Mode()
elif self._strategy == "median":
calculator = Median()
else:
raise ValueError ("Only accepts mean/mode/median")
self._ImputeValues = calculator.calculate(column_data)
def transform(self, column_index, data):
for row in data:
if row[column_index] == "nan":
row[column_index] = self._ImputeValues
return data
Result
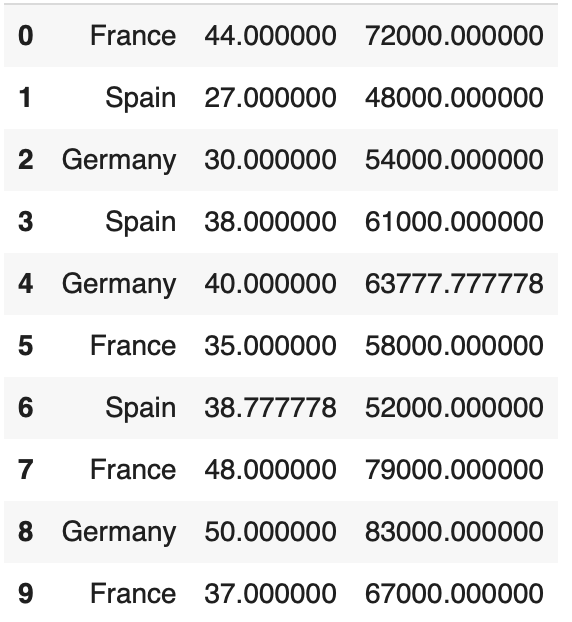
When examaning a dataset as follows:
data = [
['France', 44.0, 72000.0],
['Spain', 27.0, 48000.0],
['Germany', 30.0, 54000.0],
['Spain', 38.0, 61000.0],
['Germany', 40.0, 'nan'],
['France', 35.0, 58000.0],
['Spain', 'nan', 52000.0],
['France', 48.0, 79000.0],
['Germany', 50.0, 83000.0],
['France', 37.0, 67000.0]
]With the presented dataset, there is a simulated "none" value. With utilizing the code we can see the functionality. The code works as follows.
- Initialize the Imputer.
imputer = Imputer("mean", 0)- Establish where the Imputer should function
imputer.fit(2,data)- Performing the actual transformation
transformed_data1 = imputer.transform(2,data)the final output is as demosntrated:
The full project can be seen on my Github